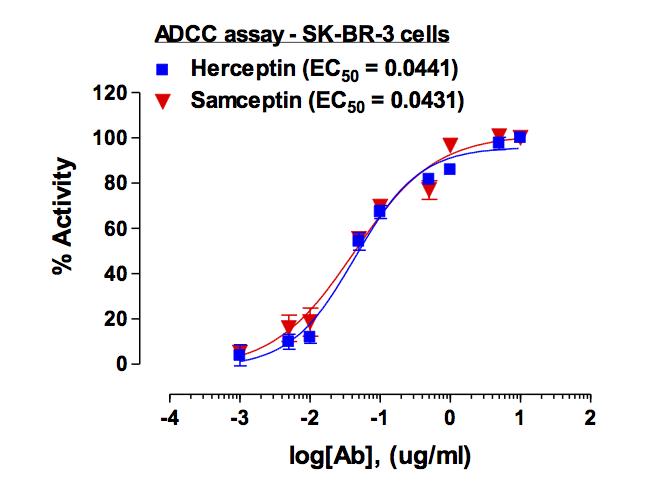
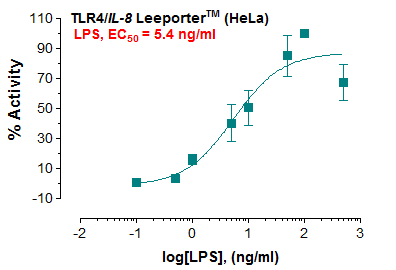
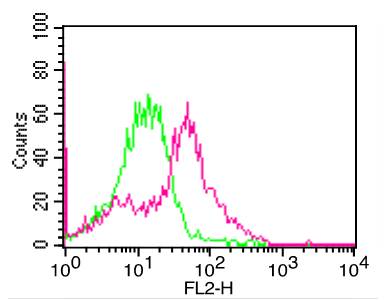
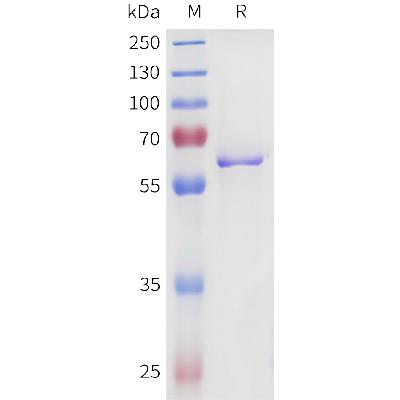
Recombinant E. coli G/U Mismatch-Specific DNA Glycosylase/Mug (C-6His)(Discontinued)
Shipping Info:
For estimated delivery dates, please contact us at [email protected]
| Amount : | 50 µg |
| Content : | Supplied as a 0.2 µm filtered solution of 20mM TrisHCl, 2.5mM beta-ME, 1mM PMSF, 50% Glycerol, pH 8.0. |
| Storage condition : | Store at -20°C, stable for 6 months after receipt. Please minimize freeze-thaw cycles. |
| AA sequence : | MVEDILAPGLRVVFCGINPGLSSAGTGFPFAHPANRFWKVIYQAGFTDRQLKPQEAQHLLDYRCGVTKLVDRPTVQANEVSKQELHAGGRKLIEKIEDYQPQALAILGKQAYEQGFSQRGAQWGKQTLTIGSTQIWVLPNPSGLSRVSLEKLVEAYRELDQALVVRGRLEHHHHHH |
Source: E.coli.
MW :19.7kD.
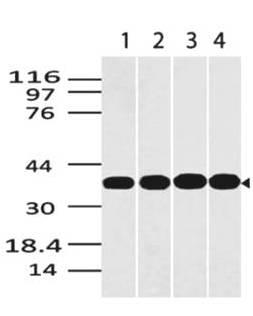
Recombinant E.coli Mug is produced by our E.coli expression system and the target gene encoding Met1-Arg168 is expressed with a 6His tag at the C-terminus. E. coli Mismatch Uracil DNA Glycosylase (Mug protein) is an 18 kDa constitutively expressed protein which belongs to the TDG/mug DNA glycosylase family. It has been proposed that the Mug protein excises 3,N4-ethenocytosine and removes the uracil base from mismatches in the order of U:G>U:A, although the biological role remains unclear. Uracil bases in DNA can arise from deamination of cytosine giving rise to increased spontaneous mutations. The enzyme Uracil-N-Glycosylase removes uracil from the DNA leaving an AP site. It is capable of hydrolyzing the carbon-nitrogen bond between the sugar-phosphate backbone of the DNA and the mispaired base. The complementary strand guanine functions in substrate recognition. It is required for DNA damage lesion repair in stationary-phase cells.
MW :19.7kD.
Recombinant E.coli Mug is produced by our E.coli expression system and the target gene encoding Met1-Arg168 is expressed with a 6His tag at the C-terminus. E. coli Mismatch Uracil DNA Glycosylase (Mug protein) is an 18 kDa constitutively expressed protein which belongs to the TDG/mug DNA glycosylase family. It has been proposed that the Mug protein excises 3,N4-ethenocytosine and removes the uracil base from mismatches in the order of U:G>U:A, although the biological role remains unclear. Uracil bases in DNA can arise from deamination of cytosine giving rise to increased spontaneous mutations. The enzyme Uracil-N-Glycosylase removes uracil from the DNA leaving an AP site. It is capable of hydrolyzing the carbon-nitrogen bond between the sugar-phosphate backbone of the DNA and the mispaired base. The complementary strand guanine functions in substrate recognition. It is required for DNA damage lesion repair in stationary-phase cells.
Endotoxin : Less than 0.1 ng/µg (1 IEU/µg) as determined by LAL test.
For Research Use Only. Not for use in diagnostic/therapeutics procedures.
| Subcellular location: | Cytoplasm |
| BioGrid: | 4262387. 156 interactions. |
|
There are currently no product reviews
|





.png)