Description
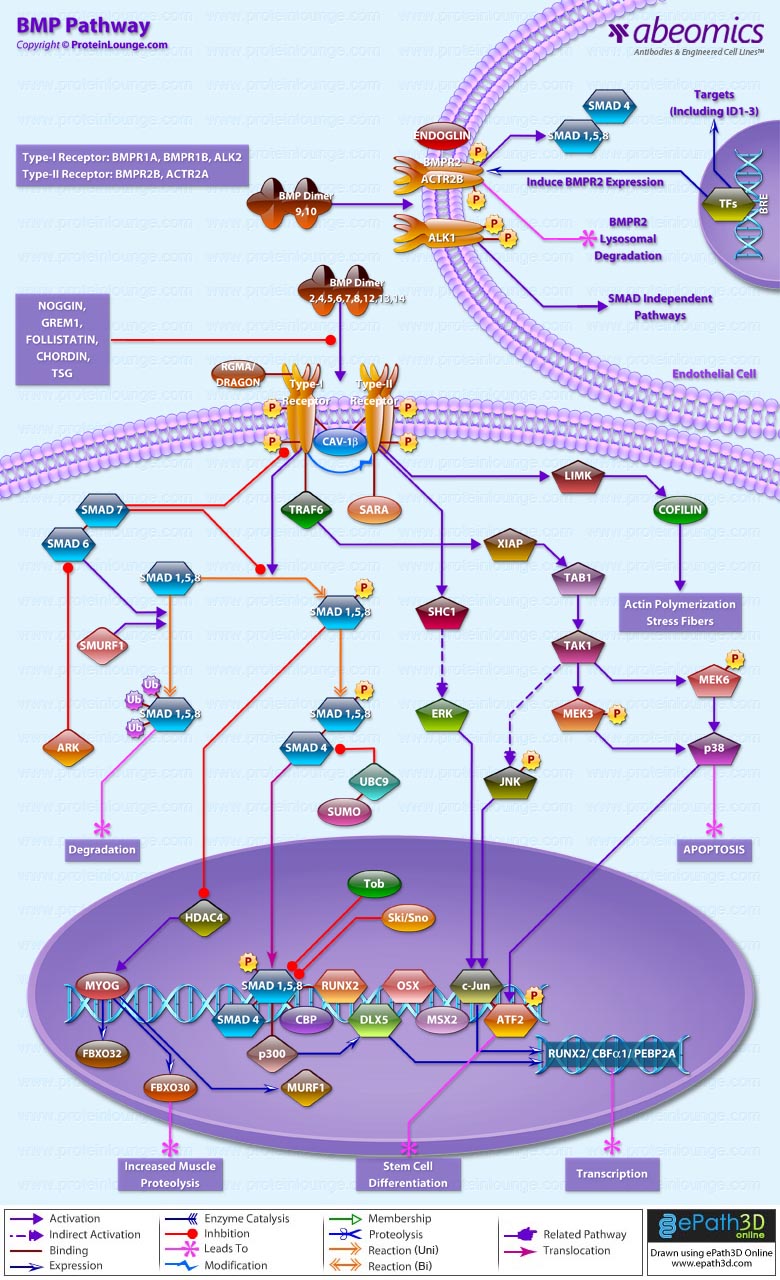
BMPs (Bone morphogenetic proteins) are the members of the transforming growth factor-beta superfamily of secreted signaling molecules [Ref.1]. Transduction of BMP signal involves two types of transmembrane serine/threonine kinase receptors: type I and type II [Ref.1]. BMP2, BMP4, BMP5,BMP6, BMP7, BMP8, BMP9, BMP10, BMP12, BMP13 and BMP14 (Bone morphogenetic protein 2, 4, 5, 6 ,7, 8, 9, 10, 12, 13 and 14) can bind to three type I receptors: BMPR1A (Bone morphogenetic protein receptor, type IA), BMPR1B (Bone morphogenetic protein receptor, type IB) and ALK-2 (Activin A receptor, type I) and two type II receptor, BMPR2A (Bone morphogenetic protein receptor type IIA) and ACTRIIA [Ref.1]. Various antagonists such as, Noggin, Tsg (twisted gastrulation), GREM1, Follistatin and Chordin bind to BMP ligands and block signaling. [Ref.2 &3]
After ligand binding, BMPs transduce their signals via two signaling pathways i.e. SMAD-dependent pathway or canonical BMP pathway and SMAD-independent or non-canonical BMP pathway [Ref.4]. In Smad dependent pathway or the canonical BMP pathway the Type II BMP receptor and type I BMP receptor is housed in a specific membrane domain of CAV1B and clathrin-coated pits at cell surface. RGMA and DRAGON acts as co-receptor of BMP signaling [Ref.1]. Meanwhile BMP ligand dimer binds to BMPRII, BMPRI is cross-phosphorylated at GS site, and recruits R-Smad (SMAD 1/5/8) to the intracellular domain of the BMPR-I and initiates signal transduction via phosphorylation [Ref.1]. Activated R-Smad then forms a heteromeric complex with C-Smad (SMAD4) [Ref.1]. This complex is translocated into the nucleus and interacts with several transcription factors such as Runx2/Cbfα1 (core binding factor alpha 1), OSX (Osterix), DLX5, and MSX2 (MSH homeobox homolog 2) [Ref.1]. These molecules mediate the transcription of related genes to induce osteogenesis [Ref.1]. However, Smad complex binds the ID1 promoter that contains two critical motifs, i.e., SBEs and Bre7 motif [Ref.1]. R-SMADs also activate transcription of target genes via complex formation with general transcriptional coactivators CBP (CREB binding protein) and p300 (E1A binding protein p300) [Ref.5 & 6]. SMAD5 and SMAD8 can also form complexes with CBP [Ref.7]. The interaction of R-SMADs with CBP, activated by CREB1 (cAMP responsive element binding protein 1), results in enhanced transcriptional activity of R-SMADs [Ref.7 & 8].
I-Smads (SMAD6/7) reside in the nucleus, migrate to the cytoplasm and can negatively regulate BMP signaling by inhibiting signal transduction at several points [Ref.1]. SMURF1 (SMAD-specific E3 ubiquitin protein ligase 1) induces ubiquitination and proteosomal degradation of R-Smad and BMPR1B [Ref.9]. I-Smad can form the complex with SMURF1 [Ref.9]. This interaction enhances degradation of R-Smad, and as well promotes SMURF1 recruitment to BMPR1B and subsequent BMPR1B ubiquitination [Ref.9]. Expression of Smad6 in denervated skeletal muscle exacerbates wasting by suppressing Smad1/5/8 phosphorylation, which in turn potentiates the HDAC4–myogenin pathway, leading to increased transcription of E3 ubiquitin ligases associated with muscle proteolysis, including MURF1, ATROGIN1, and FBXO30/MUSA1[Ref.10].SKI/SNON interacts with R-Smad, thus repressing BMP signaling [Ref.1]. TOB1, the member of anti-proliferative protein family, can directly interact with R-Smad, followed by repression of BMP/SMAD-dependent transcription and, as a consequence, osteogenesis [Ref.3]. Another way for TOB1 inhibitory action is modulating the function of I-Smad, and enhancing the interaction between SMAD6 and BMPR1B [Ref.3].
In Non-canonical Smad-independent signaling pathway, binding of ligand to the receptor leads to activation of MAPK (mitogen-activated protein kinase) pathways, including p38, ERK1/2 and JNK [Ref.1]. Type II receptor recruit TRAF6 to the cytoplasmic domain [Ref.1]. TRAF6 ubiquitylates and activates the catalytic activity of TAK1 and MAP3K7, leading to activation of the p38 and JNK (c-Jun N-terminal kinase) cascades, which regulate apoptosis and cell migration [Ref.1]. The type I receptor phosphorylates both serine and tyrosine residues in the SHCA (SHC1) adaptor, which then recruits the adaptor protein GRB2 and the GEF (Ras guanine exchange factor) SOS (son of seven less) in mammalian cells [Ref.1]. This leads to activation of the RAS-RAF-MEK-ERK, JNK, ATF2 (activating transcription factor-2), p38 (p38 MAPK), SHCA (SH2 domain-containing sequence A), and ERK (extracellular signal regulated kinase) [Ref.1]. Then, BMP-induced ATF2 activation results in stem cell differentiation [Ref.11].The signaling via BMP9/10 is specific to endothelial cells. In this pathway BMP9/10 binds to a heterotetrameric complex composed of two type I receptors (ALK1) and two type II receptors (BMPR2 and ActR2A) [Ref.12 & 13]. Endoglin is a co-receptor of this complex and enhances signaling [Ref.13]. Following ligand binding, receptors are phosphorylated and propagate signal through R-Smad phosphorylation [Ref.13]. The R-Smads then associate with Smad4 to regulate target gene transcription in the nucleus [Ref.13].
BMP signaling is critical in embryogenesis, organogenesis, adult tissue homeostasis and regulation of cardiovascular structure and function [Ref.4]. It also mediates processes important in development, such as cell proliferation, differentiation, and apoptosis [Ref.4]. Deletion of various components of the BMP pathway is embryonically lethal or presents with marked abnormalities [Ref.4]. In addition to its contributions to syndromic disorders of heart and vascular development, BMP signaling is also increasingly recognized for its influence on endocrine-like functions in postnatal cardiovascular and metabolic homeostasis [Ref.14]. Hence, it is currently a major therapeutic target skeletal trauma and disease [Ref.15].
References:
-
Rahman MS, Akhtar N, Jamil HM, Banik RS, Asaduzzaman SM.
Bone Res. 2015 Apr 14; 3:15005. doi: 10.1038/boneres.2015.5. eCollection 2015. Review.
-
Cai J, Pardali E, Sánchez-Duffhues G, ten Dijke P.
FEBS Lett. 2012 Jul 4; 586(14):1993-2002. doi: 10.1016/j.febslet.2012.04.030. Epub 2012 May 3. Review.
-
Wu M, Chen G, Li YP.
Bone Res. 2016 Apr 26; 4:16009. doi: 10.1038/boneres.2016.9. eCollection 2016. Review.
-
Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, Idowu O, Li M, Shen C, Hu A, Haydon RC, Kang R, Mok J, Lee MJ, Luu HL, Shi LL.
Genes Dis. 2014 Sep; 1(1):87-105.
-
Derynck R, Zhang YE.
Nature. 2003 Oct 9; 425(6958):577-84. Review.
-
Ross S, Hill CS.
Int J Biochem Cell Biol. 2008; 40(3):383-408. Epub 2007 Oct 7. Review.
-
Ghosh-Choudhury N, Singha PK, Woodruff K, St Clair P, Bsoul S, Werner SL, Choudhury GG.
J Biol Chem. 2006 Jul 21; 281(29):20160-70. Epub 2006 May 17.
-
Ohta Y, Nakagawa K, Imai Y, Katagiri T, Koike T, Takaoka K.
J Bone Miner Metab. 2008; 26(5):478-84. doi: 10.1007/s00774-008-0850-8. Epub 2008 Aug 30.
-
Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T.
Mol Biol Cell. 2003 Jul; 14(7):2809-17. Epub 2003 Apr 4.
-
Winbanks CE, Chen JL, Qian H, Liu Y, Bernardo BC, Beyer C, Watt KI, Thomson RE, Connor T, Turner BJ, McMullen JR, Larsson L, McGee SL, Harrison CA, Gregorevic P.
J Cell Biol. 2013 Oct 28; 203(2):345-57. doi: 10.1083/jcb.201211134. Epub 2013 Oct 21.
-
Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, Collins S.
Mol Cell Biol. 2004 Apr; 24(7):3057-67.
-
Li W, Salmon RM, Jiang H, Morrell NW.
Biochem Soc Trans. 2016 Aug 15; 44(4):1135-41. doi: 10.1042/BST20160083. Review.
-
Tillet E, Bailly S.
Front Genet. 2015 Jan 8; 5:456. doi: 10.3389/fgene.2014.00456. eCollection 2014.
-
Morrell NW, Bloch DB, ten Dijke P, Goumans MJ, Hata A, Smith J, Yu PB, Bloch KD.
Nat Rev Cardiol. 2016 Feb; 13(2):106-20. doi: 10.1038/nrcardio.2015.156. Epub 2015 Oct 13. Review.
-
Salazar VS, Gamer LW, Rosen V.
Nat Rev Endocrinol. 2016 Apr; 12(4):203-21. doi: 10.1038/nrendo.2016.12. Epub 2016 Feb 19. Review.
|